Examples for the mvn() function
Ivan Jacob Agaloos Pesigan
2020-07-17
Source:vignettes/examples/examples_mvn.Rmd
examples_mvn.Rmdlibrary(jeksterslabRdata)
Documentation
See mvn() for more details.
Examples
Single Random Data Set
Set mu and Sigma.
mu <- c(100, 100, 100) Sigma <- matrix( data = c(225, 112.50, 56.25, 112.5, 225, 112.5, 56.25, 112.50, 225), ncol = 3 )
Run the function.
X <- mvn(n = 100, mu = mu, Sigma = Sigma)
Explore the output.
str(X) #> num [1:100, 1:3] 105.8 109.3 119.7 91.5 101.9 ... #> - attr(*, "dimnames")=List of 2 #> ..$ : NULL #> ..$ : NULL psych::pairs.panels(X)
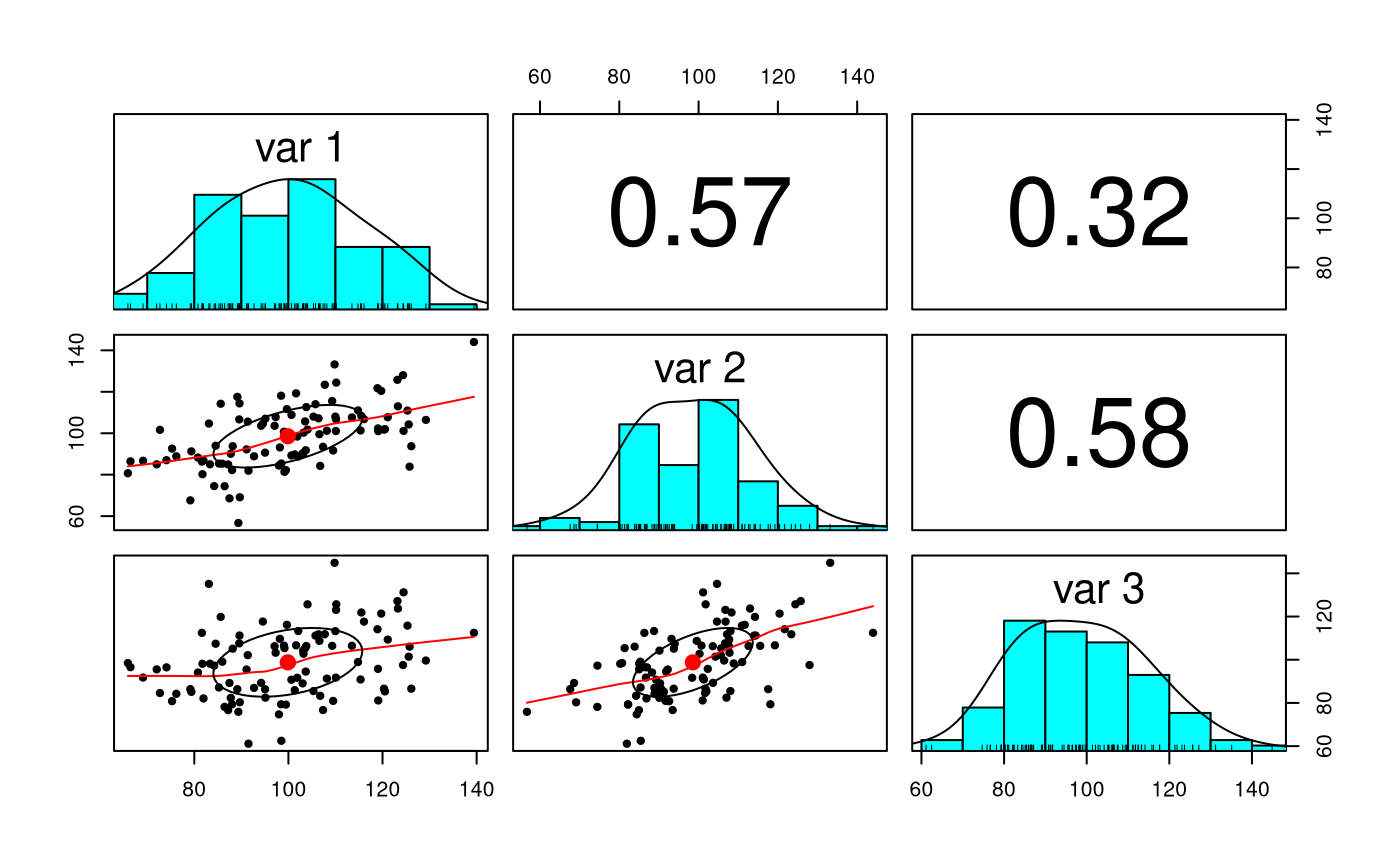
colMeans(X) #> [1] 99.87371 98.55713 98.78347 cov(X) #> [,1] [,2] [,3] #> [1,] 250.41352 135.2776 81.56604 #> [2,] 135.27762 228.4324 139.98327 #> [3,] 81.56604 139.9833 253.53888 cor(X) #> [,1] [,2] [,3] #> [1,] 1.0000000 0.5656115 0.3237116 #> [2,] 0.5656115 1.0000000 0.5816677 #> [3,] 0.3237116 0.5816677 1.0000000
Multiple Random Data Sets
Run the function.
Xstar <- mvn(n = 100, mu = mu, Sigma = Sigma, R = 1000)
Explore the output.
str(Xstar, list.len = 6) #> List of 1000 #> $ : num [1:100, 1:3] 89.7 101.8 117.6 109.5 79.5 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : NULL #> $ : num [1:100, 1:3] 99.1 82.9 91.6 108.1 96.3 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : NULL #> $ : num [1:100, 1:3] 96.4 98.6 106.2 104.9 93.6 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : NULL #> $ : num [1:100, 1:3] 73.5 92.1 103.1 68.9 100 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : NULL #> $ : num [1:100, 1:3] 112 83.4 91.5 104 83 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : NULL #> $ : num [1:100, 1:3] 104.4 98.9 99.8 125.1 95.9 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : NULL #> [list output truncated]