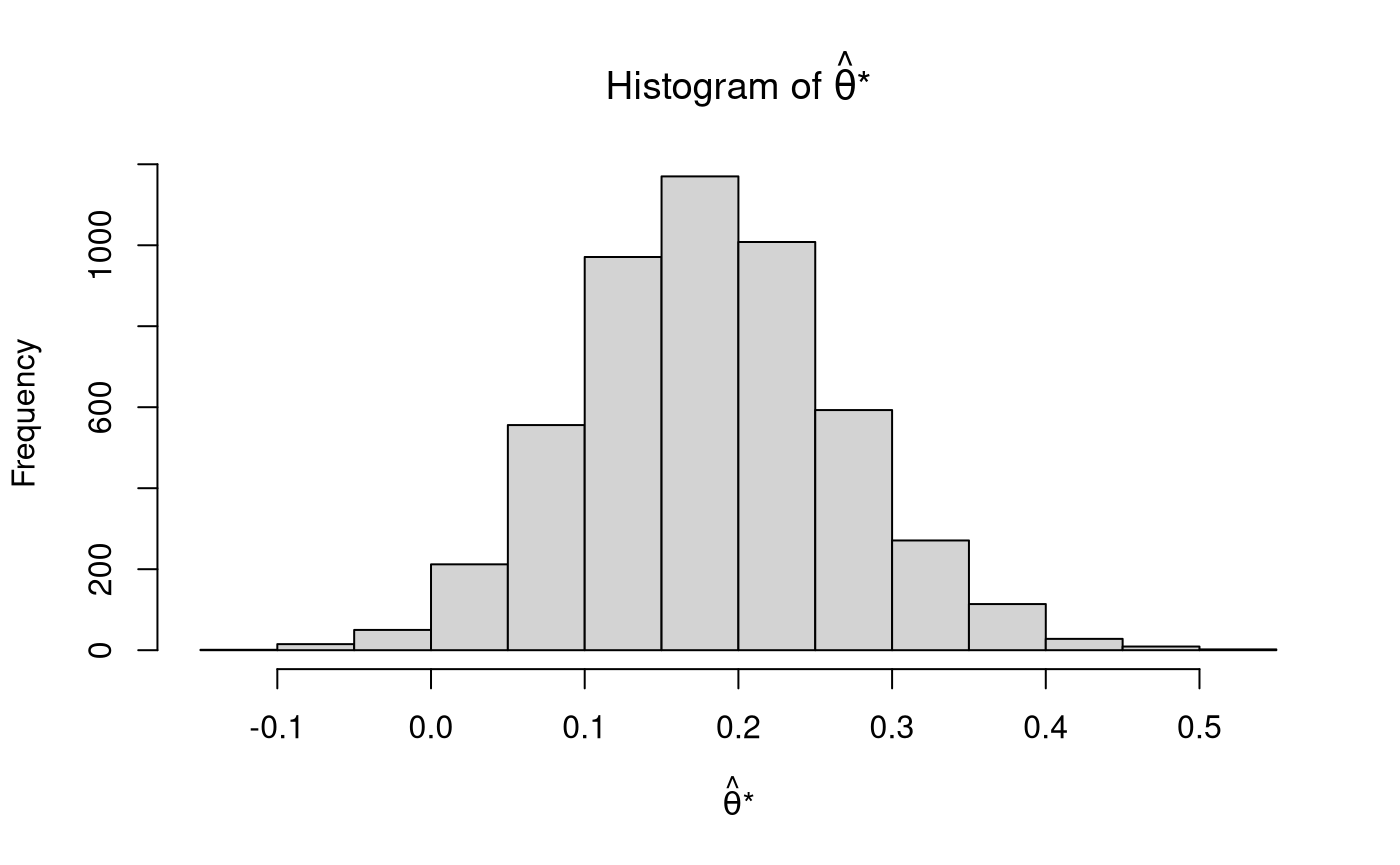
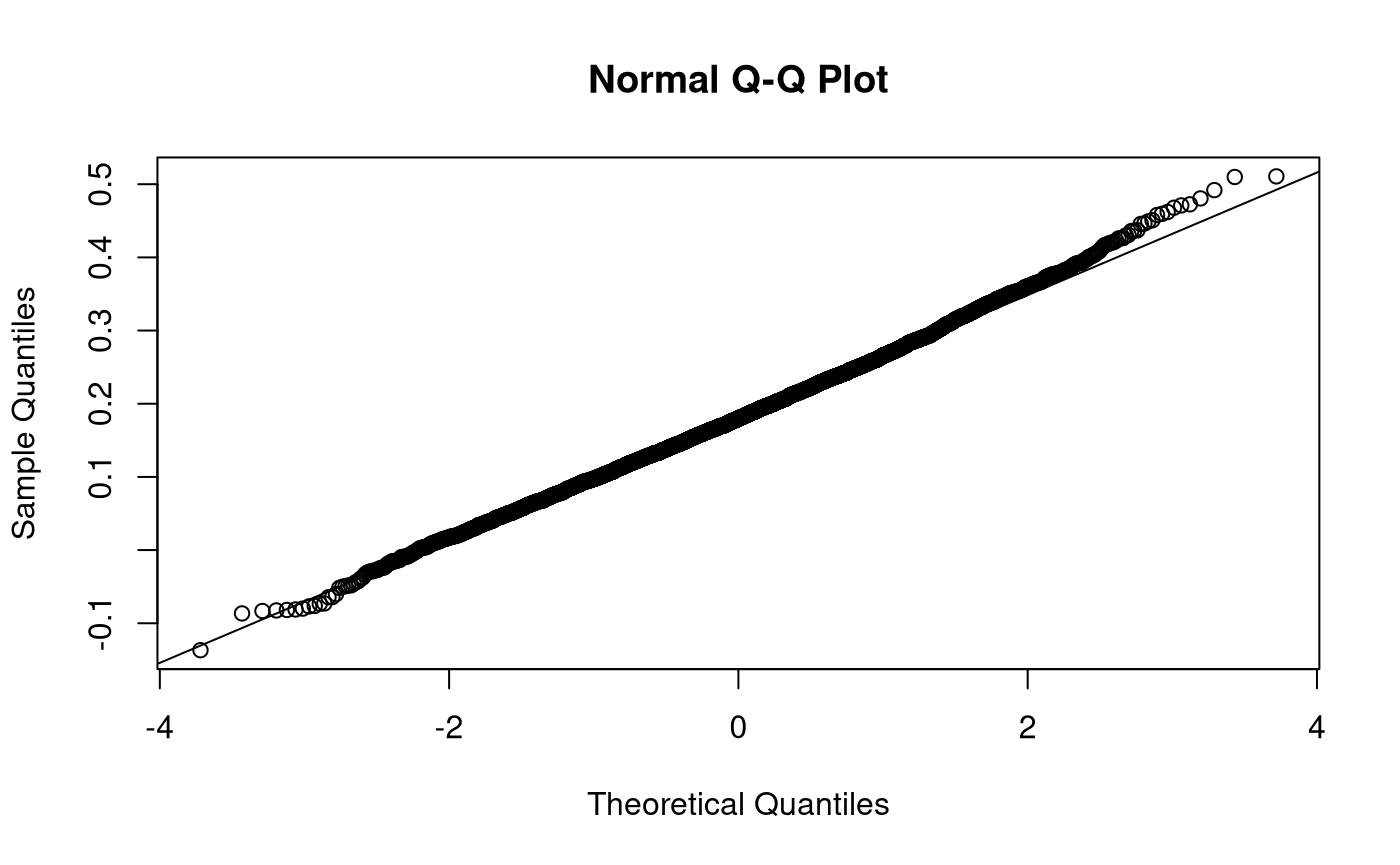
Notes: Nonparametric Bootstrapping - Mediation
Ivan Jacob Agaloos Pesigan
2020-07-16
Source:vignettes/notes/notes_nb_med.Rmd
notes_nb_med.RmdTHIS IS AN INITIAL DRAFT.
Random Variables
Population Model
\[\begin{equation} Y = \delta_Y + \tau^{\prime} X + \beta M + \varepsilon_{Y} \end{equation}\]
\[\begin{equation} M = \delta_M + \alpha X + \varepsilon_{M} \end{equation}\]
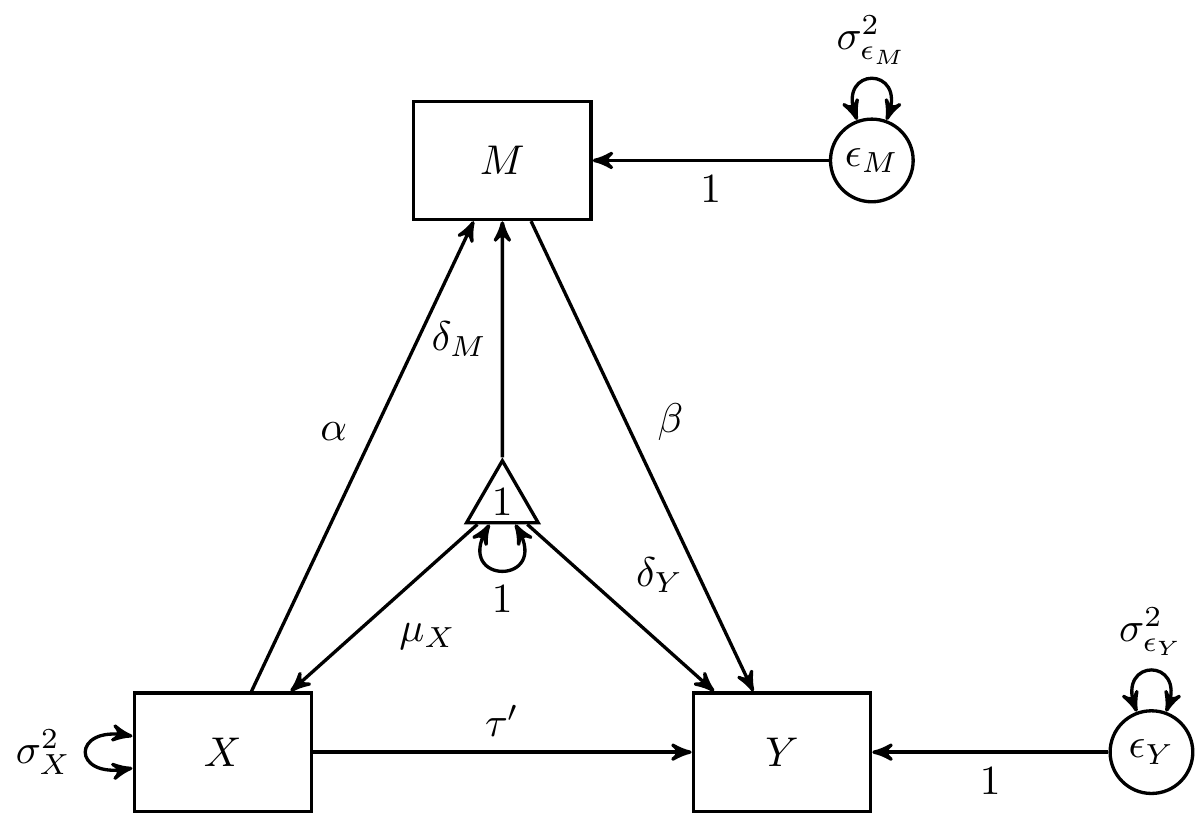
The Simple Mediation Model
Data Generating Function
\[\begin{equation} X, M, Y \sim \mathrm{MVN} \left[ \boldsymbol{\mu} \left( \boldsymbol{\theta} \right), \boldsymbol{\Sigma} \left( \boldsymbol{\theta} \right) \right] \end{equation}\]
\[\begin{equation} \boldsymbol{\mu} \left( \boldsymbol{\theta} \right) = \begin{bmatrix} \mu_X \\ \delta_M + \alpha \mu_X \\ \delta_Y + \tau^{\prime} \mu_X + \beta \delta_M + \alpha \beta \mu_X \end{bmatrix} \end{equation}\]
\[\begin{equation} \boldsymbol{\Sigma} \left( \boldsymbol{\theta} \right) = \begin{bmatrix} \sigma^{2}_{X} & \alpha \sigma_{X}^{2} & \alpha \beta \sigma_{X}^{2} + \tau^{\prime} \sigma_{X}^{2} \\ \alpha \sigma_{X}^{2} & \alpha^{2} \sigma_{X}^{2} + \sigma_{\varepsilon_{M}}^{2} & \alpha^{2} \beta \sigma_{X}^{2} + \alpha \tau^{\prime} \sigma_{X}^{2} + \beta \sigma_{\varepsilon_{M}}^{2} \\ \alpha \beta \sigma_{X}^{2} + \tau^{\prime} \sigma_{X}^{2} & \alpha^{2} \beta \sigma_{X}^{2} + \alpha \tau^{\prime} \sigma_{X}^{2} + \beta \sigma_{\varepsilon_{M}}^{2} & \beta^{2} \alpha^{2} \sigma_{X}^{2} + \beta^{2} \sigma_{\varepsilon_{M}}^{2} + 2 \alpha \beta \tau^{\prime} \sigma_{X}^{2} + \tau^{\prime} \sigma_{X}^{2} + \sigma_{\varepsilon_{Y}}^{2} \end{bmatrix} \end{equation}\]
| Variable | Description | Notation | Value |
|---|---|---|---|
muX |
Mean of \(X\). | \(\mu_X\) | 70.18000 |
deltaM |
Intercept of \(M\). | \(\delta_M\) | -20.70243 |
deltaY |
Intercept of \(Y\). | \(\delta_Y\) | -12.71288 |
| Variable | Description | Notation | Value |
|---|---|---|---|
alpha |
Regression slope of path from \(X\) to \(M\). | \(\alpha\) | 0.3385926 |
tauprime |
Regression slope of path from \(X\) to \(Y\). | \(\tau^{\prime}\) | 0.2076475 |
beta |
Regression slope of path from \(M\) to \(Y\). | \(\beta\) | 0.4510391 |
alphabeta |
Indirect effect. | \(\alpha \beta\) | 0.1527185 |
| Variable | Description | Notation | Value |
|---|---|---|---|
sigma2X |
Variance of \(X\). | \(\sigma^2_X\) | 1.2934694 |
sigma2epsilonM |
Error variance of \(\varepsilon_M\). | \(\sigma^2_{\varepsilon_{M}}\) | 0.9296694 |
sigma2epsilonY |
Error variance of \(\varepsilon_Y\). | \(\sigma^2_{\varepsilon_{Y}}\) | 0.9310601 |
| x | |
|---|---|
| X | 70.18 |
| M | 3.06 |
| Y | 3.24 |
| X | M | Y | |
|---|---|---|---|
| X | 1.2934694 | 0.4379592 | 0.4661224 |
| M | 0.4379592 | 1.0779592 | 0.5771429 |
| Y | 0.4661224 | 0.5771429 | 1.2881633 |
Sample Data
X <- as.data.frame( mvrnorm( n = n, Sigma = Sigmatheta, mu = mutheta ) ) str(X) #> 'data.frame': 50 obs. of 3 variables: #> $ X: num 69.6 72.1 68.7 68.1 70.6 ... #> $ M: num 2.78 3.07 3.14 3.92 2.97 ... #> $ Y: num 1.678 4.526 -0.146 2.965 3.084 ...
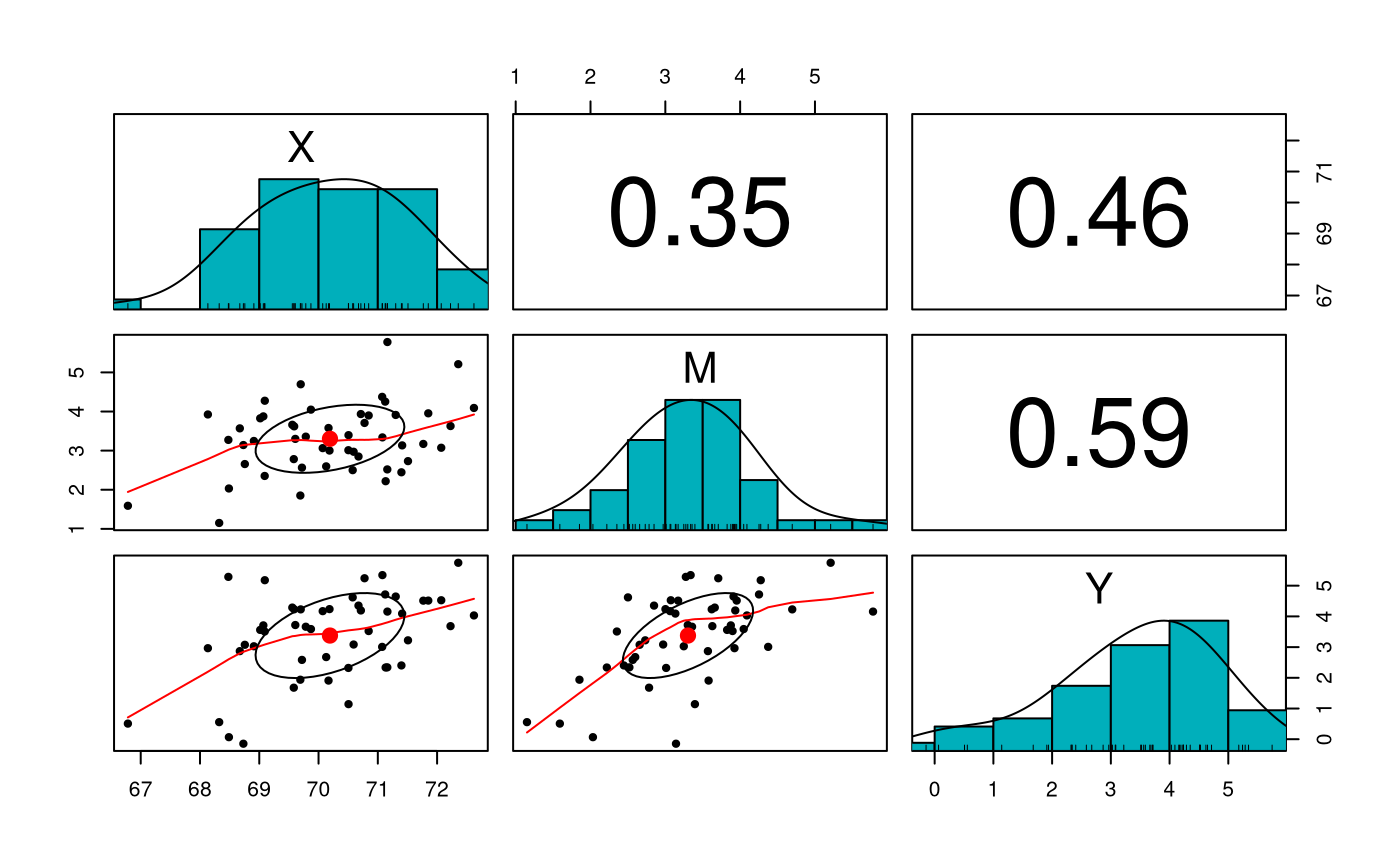
Sample Statistics
| x | |
|---|---|
| X | 70.192661 |
| M | 3.302745 |
| Y | 3.376259 |
| X | M | Y | |
|---|---|---|---|
| X | 1.5763890 | 0.3874072 | 0.8057764 |
| M | 0.3874072 | 0.7581055 | 0.7052271 |
| Y | 0.8057764 | 0.7052271 | 1.9169666 |
model_01 <- lm( M ~ X, data = X ) summary(model_01) #> #> Call: #> lm(formula = M ~ X, data = X) #> #> Residuals: #> Min 1Q Median 3Q Max #> -1.69266 -0.60955 0.07842 0.49451 2.23514 #> #> Coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) -13.9475 6.5710 -2.123 0.0390 * #> X 0.2458 0.0936 2.626 0.0116 * #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 #> #> Residual standard error: 0.8226 on 48 degrees of freedom #> Multiple R-squared: 0.1256, Adjusted R-squared: 0.1074 #> F-statistic: 6.894 on 1 and 48 DF, p-value: 0.01157 model_02 <- lm( Y ~ X + M, data = X ) summary(model_02) #> #> Call: #> lm(formula = Y ~ X + M, data = X) #> #> Residuals: #> Min 1Q Median 3Q Max #> -2.92809 -0.55738 0.02484 0.70308 2.48631 #> #> Coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) -21.8313 9.0155 -2.422 0.019368 * #> X 0.3231 0.1313 2.461 0.017593 * #> M 0.7651 0.1893 4.041 0.000196 *** #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 #> #> Residual standard error: 1.079 on 47 degrees of freedom #> Multiple R-squared: 0.4173, Adjusted R-squared: 0.3925 #> F-statistic: 16.83 on 2 and 47 DF, p-value: 3.076e-06
| Variable | Description | Notation | Value |
|---|---|---|---|
muXhat |
Estimated mean of \(X\). | \(\hat{\mu_X}\) | 70.19266 |
deltaMhat |
Estimated intercept of \(M\). | \(\hat{\delta_M}\) | -13.94753 |
deltaYhat |
Estimated intercept of \(Y\). | \(\hat{\delta_Y}\) | -21.83128 |
| Variable | Description | Notation | Value |
|---|---|---|---|
alphahat |
Estimated regression slope of path from \(X\) to \(M\). | \(\hat{\alpha}\) | 0.2457561 |
tauprimehat |
Estimated regression slope of path from \(X\) to \(Y\). | \(\hat{\tau}^{\prime}\) | 0.3231180 |
betahat |
Estimated regression slope of path from \(M\) to \(Y\). | \(\hat{\beta}\) | 0.7651294 |
alphahatbetahat |
Estimated indirect effect. | \(\hat{\alpha}\hat{\beta}\) | 0.1880352 |
| Variable | Description | Notation | Value |
|---|---|---|---|
sigma2Xhat |
Estimated variance of \(X\). | \(\hat{\sigma}^2_X\) | 1.5763890 |
sigma2epsilonMhat |
Estimated error variance of \(\varepsilon_M\). | \(\hat{\sigma}^2_{\varepsilon_{M}}\) | 0.6767082 |
sigma2epsilonYhat |
Estimated error variance of \(\varepsilon_Y\). | \(\hat{\sigma}^2_{\varepsilon_{Y}}\) | 1.1645484 |
Nonparametric Bootstrapping
Bootstrap Samples
s <- function(X) { model_01 <- lm( M ~ X, data = X ) model_02 <- lm( Y ~ X + M, data = X ) coef(model_01)[2] * coef(model_02)[3] }
Xstar <- nb( data = X, B = B )
Estimated Bootstrap Standard Error
The estimated bootstrap standard error is given by
\[\begin{equation} \widehat{\mathrm{se}}_{\mathrm{B}} \left( \hat{\theta} \right) = \sqrt{ \frac{1}{B - 1} \sum_{b = 1}^{B} \left[ \hat{\theta}^{*} \left( b \right) - \hat{\theta}^{*} \left( \cdot \right) \right]^2 } = 0.0855372 \end{equation}\]
where
\[\begin{equation} \hat{\theta}^{*} \left( \cdot \right) = \frac{1}{B} \sum_{b = 1}^{B} \hat{\theta}^{*} \left( b \right) = 0.1819124 . \end{equation}\]
Note that \(\widehat{\mathrm{se}}_{\mathrm{B}} \left( \hat{\theta} \right)\) is the standard deviation of \(\boldsymbol{\hat{\theta}^{*}}\) and \(\hat{\theta}^{*} \left( \cdot \right)\) is the mean of \(\boldsymbol{\hat{\theta}^{*}}\) .
| Variable | Description | Notation | Value |
|---|---|---|---|
B |
Number of bootstrap samples. | \(B\) | 5000.0000000 |
mean_thetahatstar |
Mean of \(B\) sample indirect effects. | \(\hat{\theta}^{*} \left( \cdot \right) = \frac{1}{B} \sum_{b = 1}^{B} \hat{\theta}^{*} \left( b \right)\) | 0.1819124 |
var_thetahatstar |
Variance of \(B\) sample indirect effects. | \(\widehat{\mathrm{Var}}_{\mathrm{B}} \left( \hat{\theta} \right) = \frac{1}{B - 1} \sum_{b = 1}^{B} \left[ \hat{\theta}^{*} \left( b \right) - \hat{\theta}^{*} \left( \cdot \right) \right]^2\) | 0.0073166 |
sd_thetahatstar |
Standard deviation of \(B\) sample indirect effects. | \(\widehat{\mathrm{se}}_{\mathrm{B}} \left( \hat{\theta} \right) = \sqrt{ \frac{1}{B - 1} \sum_{b = 1}^{B} \left[ \hat{\theta}^{*} \left( b \right) - \hat{\theta}^{*} \left( \cdot \right) \right]^2 }\) | 0.0855372 |
Confidence Intervals
Confidence intervals can be constructed around \(\hat{\theta}\) . The functions pc(), bc(), and bca() from the jeksterslabRboot package can be used to construct confidence intervals using the default alphas of 0.001, 0.01, and 0.05. The confidence intervals can also be evaluated. Since we know the population parameter theta \(\left(\theta = \mu = 0.1527185 \right)\), we can check if our confidence intervals contain the population parameter.
See documentation for pc(), bc(), and bca() from the jeksterslabRboot package on how confidence intervals are constructed.
See documentation for zero_hit(), theta_hit(), len(), and shape() from the jeksterslabRboot package on how confidence intervals are evaluated.
pc_out <- pc( thetahatstar = thetahatstar, thetahat = thetahat, eval = TRUE, theta = theta ) bc_out <- bc( thetahatstar = thetahatstar, thetahat = thetahat, eval = TRUE, theta = theta ) bca_out <- bca( thetahatstar = thetahatstar, thetahat = thetahat, data = X, fitFUN = s, eval = TRUE, theta = theta )
| statistic | p | se | ci_0.05 | ci_0.5 | ci_2.5 | ci_97.5 | ci_99.5 | ci_99.95 | zero_hit_0.001 | zero_hit_0.01 | zero_hit_0.05 | theta_hit_0.001 | theta_hit_0.01 | theta_hit_0.05 | length_0.001 | length_0.01 | length_0.05 | shape_0.001 | shape_0.01 | shape_0.05 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pc | NA | NA | 0.0855372 | -0.0828897 | -0.0312716 | 0.0193249 | 0.3558904 | 0.4195786 | 0.4862799 | 1 | 1 | 0 | 1 | 1 | 1 | 0.5691697 | 0.4508501 | 0.3365655 | 1.100839 | 1.055796 | 0.9949315 |
| bc | NA | NA | 0.0855372 | -0.0813504 | -0.0152843 | 0.0348956 | 0.3740522 | 0.4368202 | 0.5048616 | 1 | 1 | 0 | 1 | 1 | 1 | 0.5862121 | 0.4521045 | 0.3391567 | 1.176107 | 1.223616 | 1.2146883 |
| bca | NA | NA | 0.0855372 | -0.0544361 | 0.0031244 | 0.0447128 | 0.3871218 | 0.4682649 | 0.5105386 | 1 | 0 | 0 | 1 | 1 | 1 | 0.5649747 | 0.4651405 | 0.3424091 | 1.330068 | 1.515485 | 1.3890815 |
References
- Bollen, K. A., & Stine, R., (1990). Direct and indirect effects: Classical and bootstrap estimates of variability. Sociological Methodology, 20, 115-40. https://doi.org/10.2307/271084
- Efron, B., & Tibshirani, R. J. (1993). An introduction to the bootstrap. New York, N.Y: Chapman & Hall.
- MacKinnon, D. P. (2008). Introduction to statistical mediation analysis. New York: Lawrence Erlbaum Associates.
- Shrout, P. E., & Bolger, N. (2002). Mediation in experimental and nonexperimental studies: New procedures and recommendations. Psychological Methods, 7, 422-445. https://doi.org/10.1037/1082-989X.7.4.422
- Wikipedia: Bootstrapping (statistics)