Notes: Model of the Stability of Alienation
Ivan Jacob Agaloos Pesigan
2020-12-31
Source:vignettes/notes/notes_alienation.Rmd
notes_alienation.RmdLoad Data
data(
alienation,
package = "jeksterslabRdatarepo"
)Data Table
Variance-Covariance Matrix
| anomia67 | powerless67 | anomia71 | powerless71 | education | sei | |
|---|---|---|---|---|---|---|
| anomia67 | 11.834 | 6.947 | 6.819 | 4.783 | -3.839 | -21.899 |
| powerless67 | 6.947 | 9.364 | 5.091 | 5.028 | -3.889 | -18.831 |
| anomia71 | 6.819 | 5.091 | 12.532 | 7.495 | -3.841 | -21.748 |
| powerless71 | 4.783 | 5.028 | 7.495 | 9.986 | -3.625 | -18.775 |
| education | -3.839 | -3.889 | -3.841 | -3.625 | 9.610 | 35.522 |
| sei | -21.899 | -18.831 | -21.748 | -18.775 | 35.522 | 450.288 |
Model
Saturated Model
# model specification
model_cov <- "
# covariances
# col 1
anomia67 ~~ anomia67
anomia71 ~~ anomia67
powerless67 ~~ anomia67
powerless71 ~~ anomia67
education ~~ anomia67
sei ~~ anomia67
# col 2
anomia71 ~~ anomia71
powerless67 ~~ anomia71
powerless71 ~~ anomia71
education ~~ anomia71
sei ~~ anomia71
# col 3
powerless67 ~~ powerless67
powerless71 ~~ powerless67
education ~~ powerless67
sei ~~ powerless67
# col 4
powerless71 ~~ powerless71
education ~~ powerless71
sei ~~ powerless71
# col 5
education ~~ education
sei ~~ education
# col 6
sei ~~ sei
"
# model fitting
fit_cov <- sem(
model_cov,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_cov, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 146 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 21
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 0.000
#> Degrees of freedom 0
#>
#> Model Test Baseline Model:
#>
#> Test statistic 2133.722
#> Degrees of freedom 15
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 1.000
#> Tucker-Lewis Index (TLI) 1.000
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -15210.906
#> Loglikelihood unrestricted model (H1) -15210.906
#>
#> Akaike (AIC) 30463.813
#> Bayesian (BIC) 30565.397
#> Sample-size adjusted Bayesian (BIC) 30498.703
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.000
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.000
#> P-value RMSEA <= 0.05 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> anomia67 ~~
#> anomia71 6.812 0.457 14.915 0.000 6.812 0.560
#> powerless67 6.940 0.413 16.815 0.000 6.940 0.660
#> powerless71 4.778 0.389 12.295 0.000 4.778 0.440
#> education -3.835 0.371 -10.340 0.000 -3.835 -0.360
#> sei -21.876 2.494 -8.772 0.000 -21.876 -0.300
#> anomia71 ~~
#> powerless67 5.086 0.392 12.985 0.000 5.086 0.470
#> powerless71 7.487 0.441 16.992 0.000 7.487 0.670
#> education -3.837 0.380 -10.085 0.000 -3.837 -0.350
#> sei -21.725 2.559 -8.490 0.000 -21.725 -0.290
#> powerless67 ~~
#> powerless71 5.023 0.357 14.084 0.000 5.023 0.520
#> education -3.885 0.335 -11.580 0.000 -3.885 -0.410
#> sei -18.811 2.212 -8.503 0.000 -18.811 -0.290
#> powerless71 ~~
#> education -3.621 0.342 -10.595 0.000 -3.621 -0.370
#> sei -18.755 2.279 -8.231 0.000 -18.755 -0.280
#> education ~~
#> sei 35.484 2.446 14.506 0.000 35.484 0.540
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> anomia67 11.821 0.548 21.587 0.000 11.821 1.000
#> anomia71 12.519 0.580 21.587 0.000 12.519 1.000
#> powerless67 9.354 0.433 21.587 0.000 9.354 1.000
#> powerless71 9.975 0.462 21.587 0.000 9.975 1.000
#> education 9.600 0.445 21.587 0.000 9.600 1.000
#> sei 449.805 20.837 21.587 0.000 449.805 1.000
semPaths(fit_cov, what = "path", whatLabels = "est", style = "ram", layout = "circle")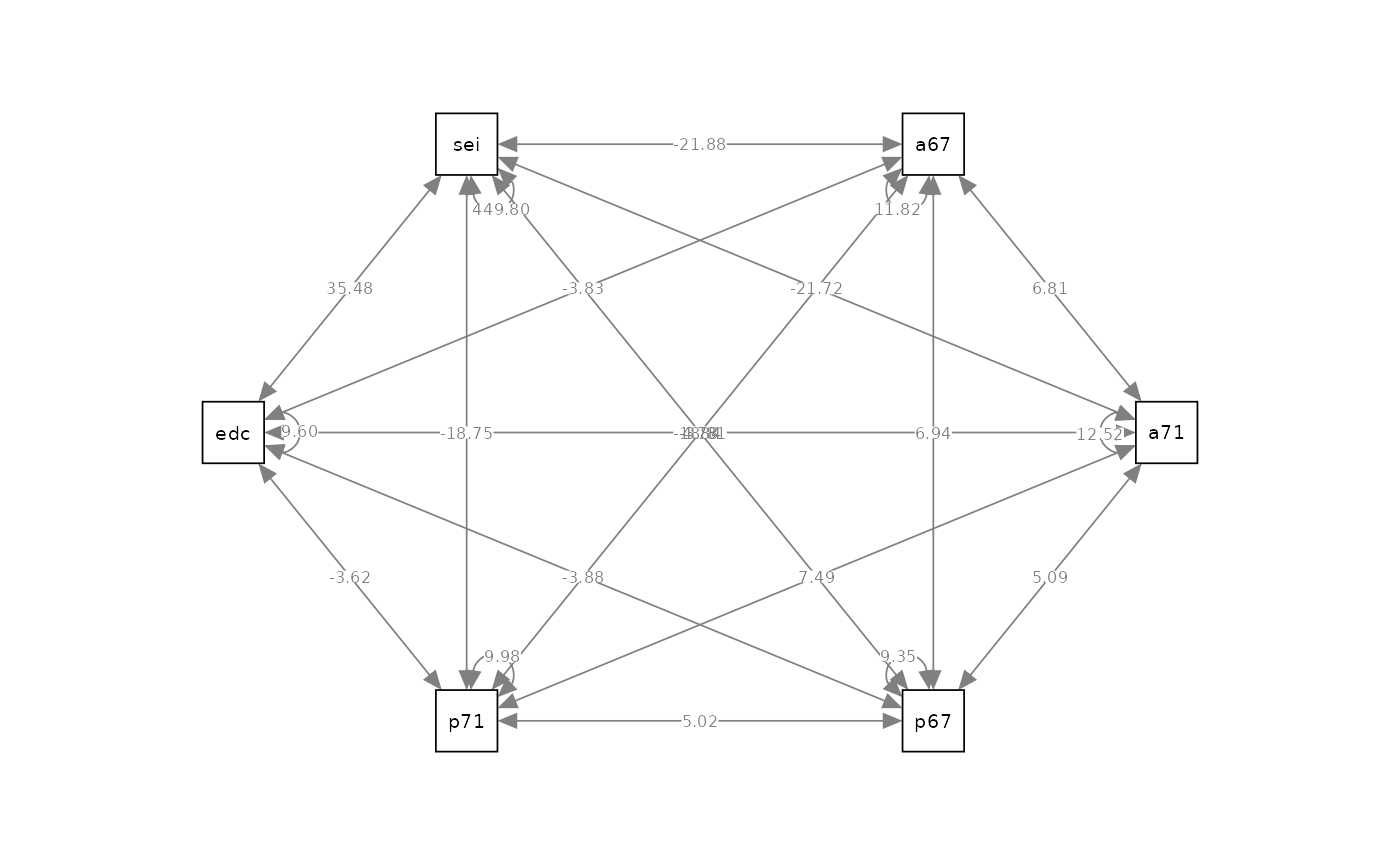
Linear Regression Model
# model specification
model_regression <- "
# regression
powerless71 ~ powerless67 + education
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# variances of regressors
powerless67 ~~ powerless67
education ~~ education
# covariance of regressors
powerless67 ~~ education
# residual variance
powerless71 ~~ powerless71
"
# model fitting
fit_regression <- sem(
model_regression,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_regression, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 21 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 6
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 0.000
#> Degrees of freedom 0
#>
#> Model Test Baseline Model:
#>
#> Test statistic 503.835
#> Degrees of freedom 3
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 1.000
#> Tucker-Lewis Index (TLI) 1.000
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -6883.135
#> Loglikelihood unrestricted model (H1) -6883.135
#>
#> Akaike (AIC) 13778.270
#> Bayesian (BIC) 13807.294
#> Sample-size adjusted Bayesian (BIC) 13788.238
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.000
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.000
#> P-value RMSEA <= 0.05 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> powerless71 ~
#> powerless67 0.457 0.031 14.731 0.000 0.457 0.443
#> education -0.192 0.031 -6.276 0.000 -0.192 -0.189
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> powerless67 ~~
#> education -3.885 0.335 -11.580 0.000 -3.885 -0.410
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> powerless67 9.354 0.433 21.587 0.000 9.354 1.000
#> education 9.600 0.445 21.587 0.000 9.600 1.000
#> .powerless71 6.983 0.323 21.587 0.000 6.983 0.700
semPaths(fit_regression, what = "path", whatLabels = "est", style = "ram")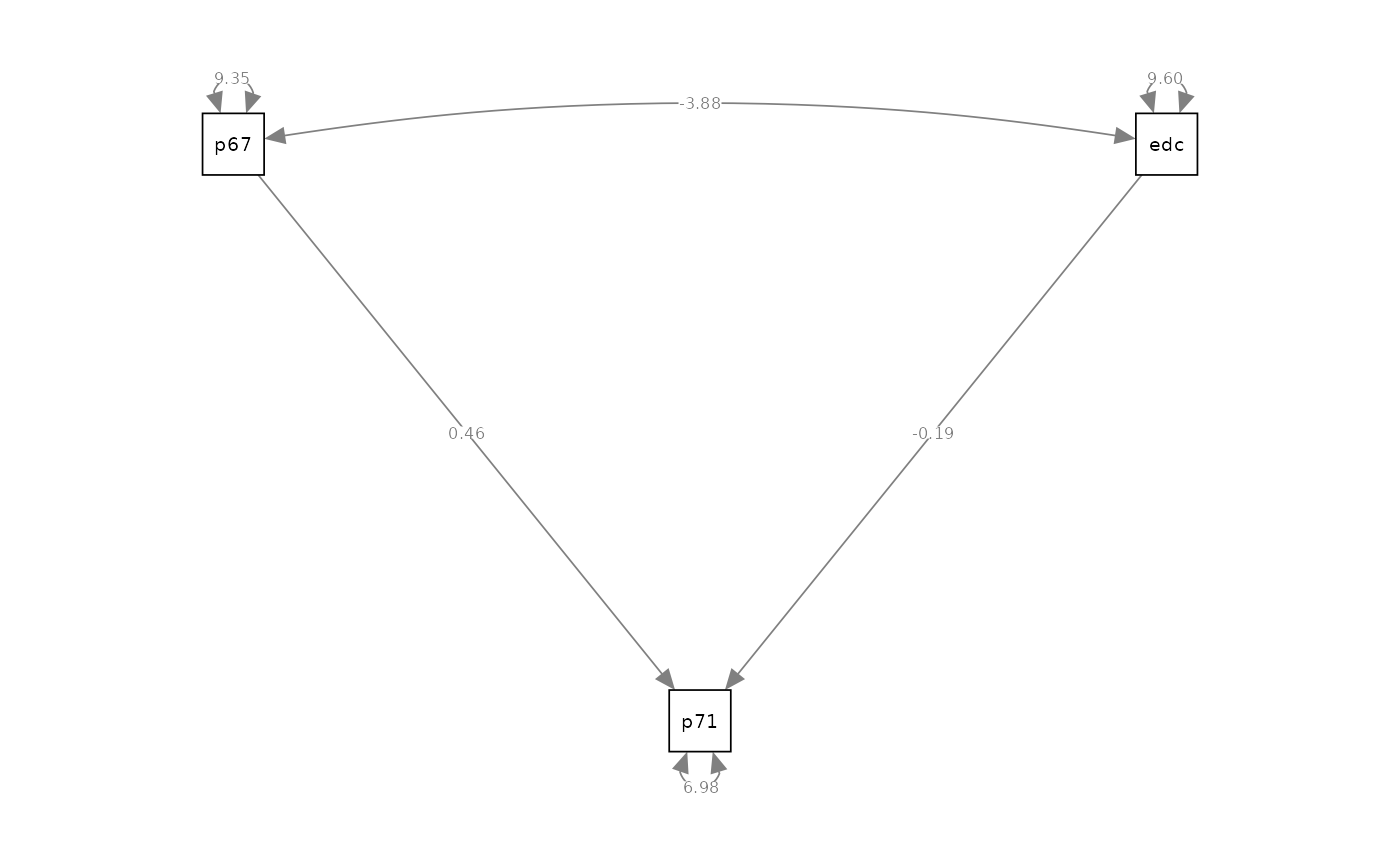
Two Equation Path Model
# model specification
model_path <- "
# regression
anomia71 ~ anomia67 + powerless67
powerless71 ~ anomia67 + powerless67
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# variances of regressors
anomia67 ~~ anomia67
powerless67 ~~ powerless67
# covariance of regressors
anomia67 ~~ powerless67
# residual variances
anomia71 ~~ anomia71
powerless71 ~~ powerless71
"
# model fitting
fit_path <- sem(
model_path,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_path, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 36 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 10
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 0.000
#> Degrees of freedom 0
#>
#> Model Test Baseline Model:
#>
#> Test statistic 1565.624
#> Degrees of freedom 6
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 1.000
#> Tucker-Lewis Index (TLI) 1.000
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -8949.380
#> Loglikelihood unrestricted model (H1) -8949.380
#>
#> Akaike (AIC) 17918.761
#> Bayesian (BIC) 17967.134
#> Sample-size adjusted Bayesian (BIC) 17935.375
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.000
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.000
#> P-value RMSEA <= 0.05 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> anomia71 ~
#> anomia67 0.455 0.037 12.413 0.000 0.455 0.443
#> powerless67 0.206 0.041 4.991 0.000 0.206 0.178
#> powerless71 ~
#> anomia67 0.158 0.034 4.660 0.000 0.158 0.172
#> powerless67 0.420 0.038 11.048 0.000 0.420 0.407
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> anomia67 ~~
#> powerless67 6.940 0.413 16.815 0.000 6.940 0.660
#> .anomia71 ~~
#> .powerless71 4.277 0.289 14.802 0.000 4.277 0.554
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> anomia67 11.821 0.548 21.587 0.000 11.821 1.000
#> powerless67 9.354 0.433 21.587 0.000 9.354 1.000
#> .anomia71 8.370 0.388 21.587 0.000 8.370 0.669
#> .powerless71 7.113 0.329 21.587 0.000 7.113 0.713
semPaths(object = fit_path, what = "path", whatLabels = "est", style = "ram")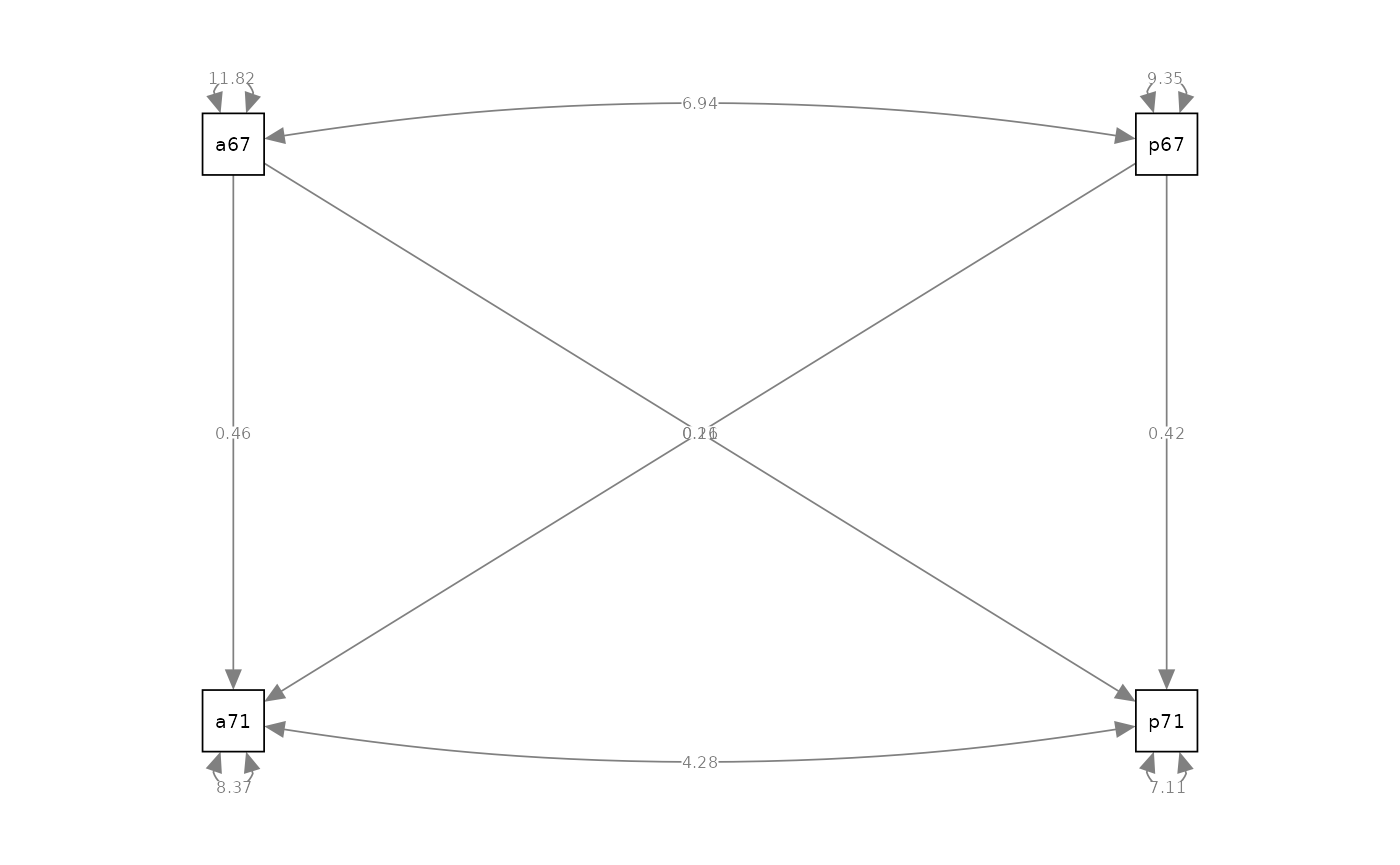
Factor Model
One-Factor Model with Two Indicators
# model specification
model_cfa_1 <- "
# measurement model
alienation =~ 1 * anomia67 + powerless67
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# residual variances
anomia67 ~~ anomia67
powerless67 ~~ powerless67
alienation ~~ alienation
"
# model fitting
fit_cfa_1 <- sem(
model_cfa_1,
sample.cov = alienation,
sample.nobs = 932
)
#> Warning in lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, : lavaan WARNING:
#> Could not compute standard errors! The information matrix could
#> not be inverted. This may be a symptom that the model is not
#> identified.
#> Warning in lav_object_post_check(object): lavaan WARNING: some estimated ov
#> variances are negative
# results
summary(fit_cfa_1, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 23 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 4
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic NA
#> Degrees of freedom -1
#> P-value (Unknown) NA
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) NA
#> Tucker-Lewis Index (TLI) NA
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -4571.282
#> Loglikelihood unrestricted model (H1) -4571.282
#>
#> Akaike (AIC) 9150.564
#> Bayesian (BIC) 9169.913
#> Sample-size adjusted Bayesian (BIC) 9157.209
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA NA
#> 90 Percent confidence interval - lower NA
#> 90 Percent confidence interval - upper NA
#> P-value RMSEA <= 0.05 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Latent Variables:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation =~
#> anomia67 1.000 1.905 0.554
#> powerless67 1.912 NA 3.643 1.191
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .anomia67 8.193 NA 8.193 0.693
#> .powerless67 -3.917 NA -3.917 -0.419
#> alienation 3.629 NA 1.000 1.000
semPaths(fit_cfa_1, what = "path", whatLabels = "est", style = "ram")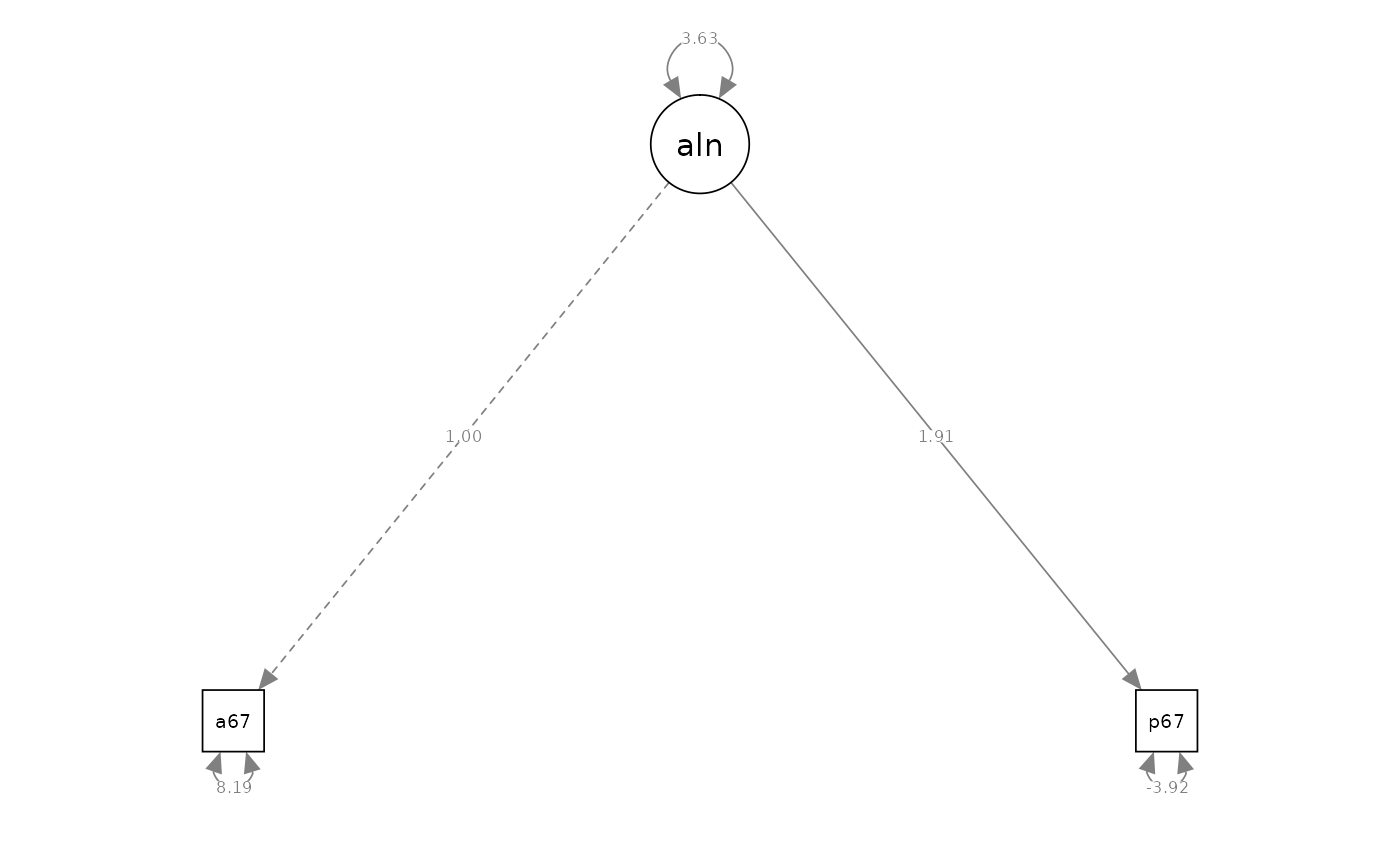
One-Factor Model with Four Indicators
# model specification
model_cfa_2 <- "
# measurement model
alienation =~ 1 * anomia67 + powerless67 + anomia71 + powerless71
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# residual variances
anomia67 ~~ anomia67
powerless67 ~~ powerless67
anomia71 ~~ anomia71
powerless71 ~~ powerless71
alienation ~~ alienation
"
# model fitting
fit_cfa_2 <- sem(
model_cfa_2,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_cfa_2, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 30 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 8
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 220.877
#> Degrees of freedom 2
#> P-value (Chi-square) 0.000
#>
#> Model Test Baseline Model:
#>
#> Test statistic 1565.624
#> Degrees of freedom 6
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 0.860
#> Tucker-Lewis Index (TLI) 0.579
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -9059.819
#> Loglikelihood unrestricted model (H1) -8949.380
#>
#> Akaike (AIC) 18135.638
#> Bayesian (BIC) 18174.336
#> Sample-size adjusted Bayesian (BIC) 18148.929
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.343
#> 90 Percent confidence interval - lower 0.305
#> 90 Percent confidence interval - upper 0.382
#> P-value RMSEA <= 0.05 0.000
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.067
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Latent Variables:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation =~
#> anomia67 1.000 2.537 0.738
#> powerless67 0.874 0.044 19.779 0.000 2.218 0.725
#> anomia71 1.080 0.052 20.806 0.000 2.741 0.775
#> powerless71 0.918 0.046 20.053 0.000 2.329 0.737
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .anomia67 5.384 0.333 16.180 0.000 5.384 0.455
#> .powerless67 4.433 0.267 16.580 0.000 4.433 0.474
#> .anomia71 5.006 0.339 14.774 0.000 5.006 0.400
#> .powerless71 4.552 0.281 16.199 0.000 4.552 0.456
#> alienation 6.437 0.535 12.032 0.000 1.000 1.000
semPaths(fit_cfa_2, what = "path", whatLabels = "est", style = "ram")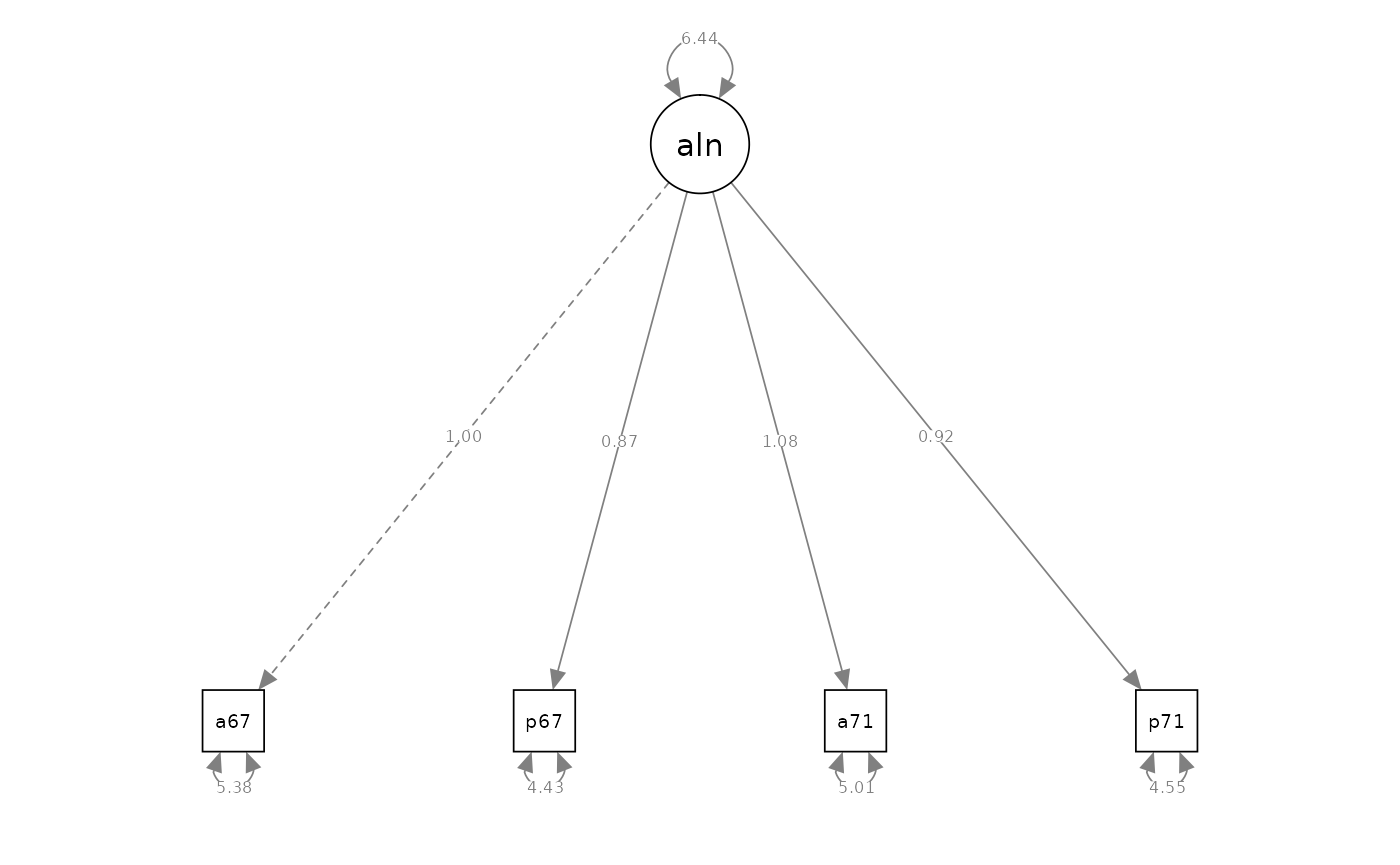
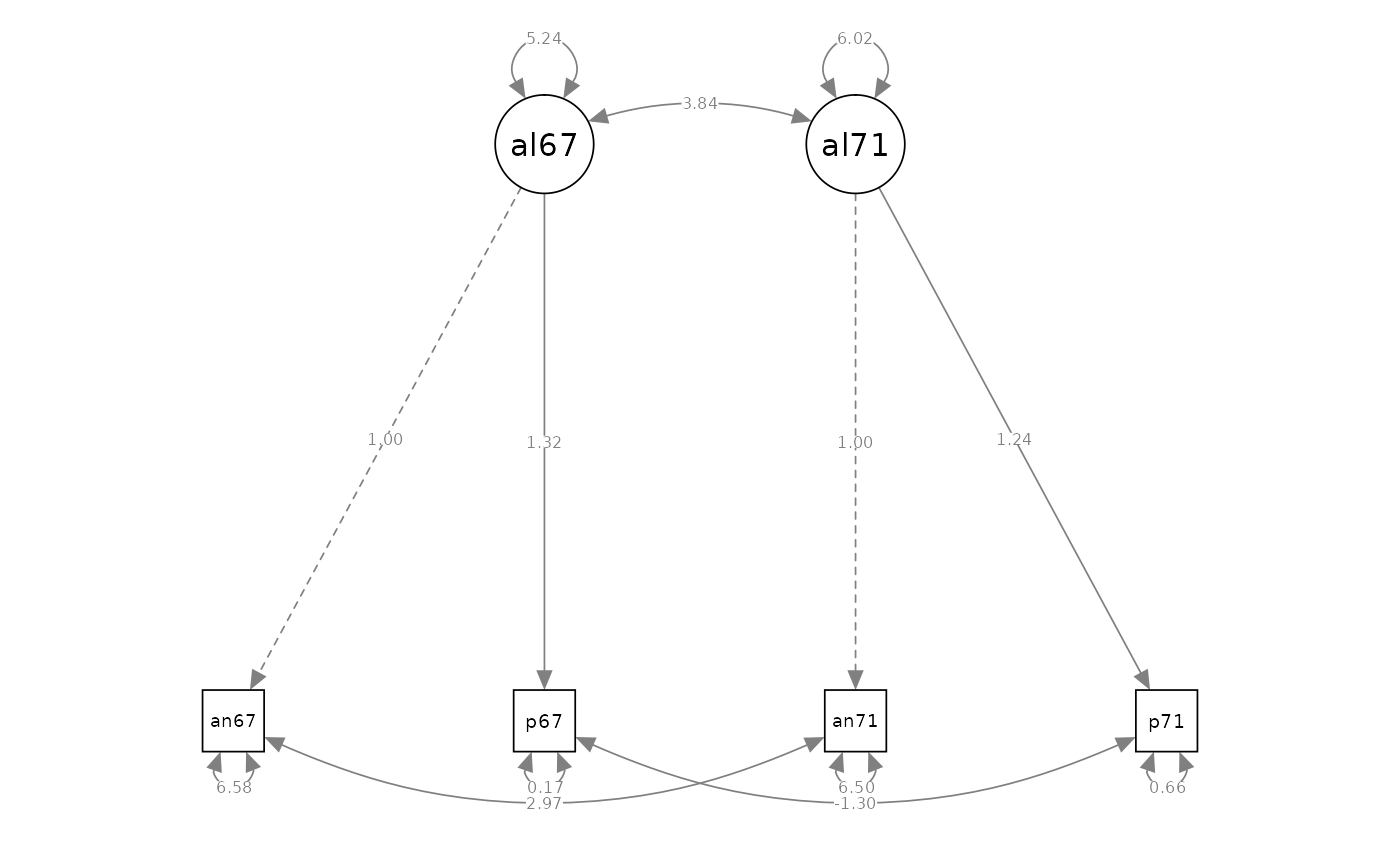
Two-Factor Model - Parameterization 1
# model specification
model_cfa_3 <- "
# measurement model
alienation67 =~ 1 * anomia67 + powerless67
alienation71 =~ 1 * anomia71 + powerless71
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# covariance of factors
alienation67 ~~ alienation71
# variances of factors
alienation67 ~~ alienation67
alienation71 ~~ alienation71
# residual variances
anomia67 ~~ anomia67
powerless67 ~~ powerless67
anomia71 ~~ anomia71
powerless71 ~~ powerless71
"
# model fitting
fit_cfa_3 <- sem(
model_cfa_3,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_cfa_3, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 48 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 9
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 61.173
#> Degrees of freedom 1
#> P-value (Chi-square) 0.000
#>
#> Model Test Baseline Model:
#>
#> Test statistic 1565.624
#> Degrees of freedom 6
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 0.961
#> Tucker-Lewis Index (TLI) 0.769
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -8979.967
#> Loglikelihood unrestricted model (H1) -8949.380
#>
#> Akaike (AIC) 17977.933
#> Bayesian (BIC) 18021.469
#> Sample-size adjusted Bayesian (BIC) 17992.886
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.254
#> 90 Percent confidence interval - lower 0.202
#> 90 Percent confidence interval - upper 0.310
#> P-value RMSEA <= 0.05 0.000
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.027
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Latent Variables:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 =~
#> anomia67 1.000 2.862 0.832
#> powerless67 0.847 0.042 20.169 0.000 2.425 0.793
#> alienation71 =~
#> anomia71 1.000 3.030 0.856
#> powerless71 0.815 0.040 20.354 0.000 2.471 0.782
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 ~~
#> alienation71 6.465 0.449 14.399 0.000 0.745 0.745
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 8.192 0.617 13.272 0.000 1.000 1.000
#> alienation71 9.183 0.668 13.742 0.000 1.000 1.000
#> .anomia67 3.629 0.371 9.784 0.000 3.629 0.307
#> .powerless67 3.475 0.287 12.119 0.000 3.475 0.372
#> .anomia71 3.335 0.398 8.390 0.000 3.335 0.266
#> .powerless71 3.871 0.302 12.803 0.000 3.871 0.388
semPaths(fit_cfa_3, what = "path", whatLabels = "est", style = "ram")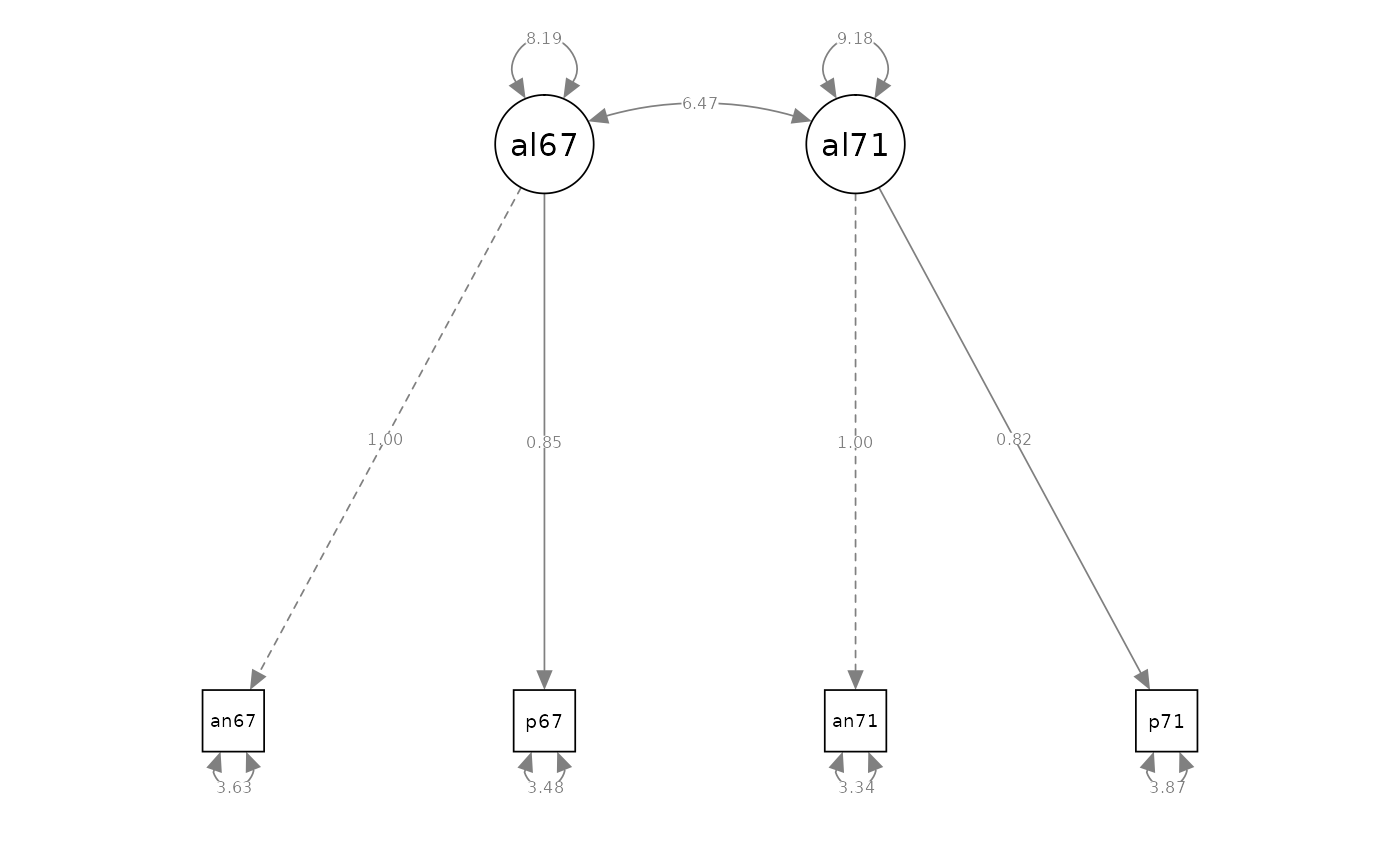
Two-Factor Model - Parameterization 2
# model specification
model_cfa_4 <- "
# measurement model
alienation67 =~ NA * anomia67 + powerless67
alienation71 =~ NA * anomia71 + powerless71
# variances of factors
alienation67 ~~ 1 * alienation67
alienation71 ~~ 1 * alienation71
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# covariance of factors
alienation67 ~~ alienation71
# measurement error variances
anomia67 ~~ anomia67
powerless67 ~~ powerless67
anomia71 ~~ anomia71
powerless71 ~~ powerless71
"
# model fitting
fit_cfa_4 <- sem(
model_cfa_4,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_cfa_4, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 19 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 9
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 61.173
#> Degrees of freedom 1
#> P-value (Chi-square) 0.000
#>
#> Model Test Baseline Model:
#>
#> Test statistic 1565.624
#> Degrees of freedom 6
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 0.961
#> Tucker-Lewis Index (TLI) 0.769
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -8979.967
#> Loglikelihood unrestricted model (H1) -8949.380
#>
#> Akaike (AIC) 17977.933
#> Bayesian (BIC) 18021.469
#> Sample-size adjusted Bayesian (BIC) 17992.886
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.254
#> 90 Percent confidence interval - lower 0.202
#> 90 Percent confidence interval - upper 0.310
#> P-value RMSEA <= 0.05 0.000
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.027
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Latent Variables:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 =~
#> anomia67 2.862 0.108 26.544 0.000 2.862 0.832
#> powerless67 2.425 0.096 25.172 0.000 2.425 0.793
#> alienation71 =~
#> anomia71 3.030 0.110 27.484 0.000 3.030 0.856
#> powerless71 2.471 0.099 24.886 0.000 2.471 0.782
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 ~~
#> alienation71 0.745 0.024 30.522 0.000 0.745 0.745
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 1.000 1.000 1.000
#> alienation71 1.000 1.000 1.000
#> .anomia67 3.629 0.371 9.784 0.000 3.629 0.307
#> .powerless67 3.475 0.287 12.119 0.000 3.475 0.372
#> .anomia71 3.335 0.398 8.390 0.000 3.335 0.266
#> .powerless71 3.871 0.302 12.803 0.000 3.871 0.388
semPaths(fit_cfa_4, what = "path", whatLabels = "est", style = "ram")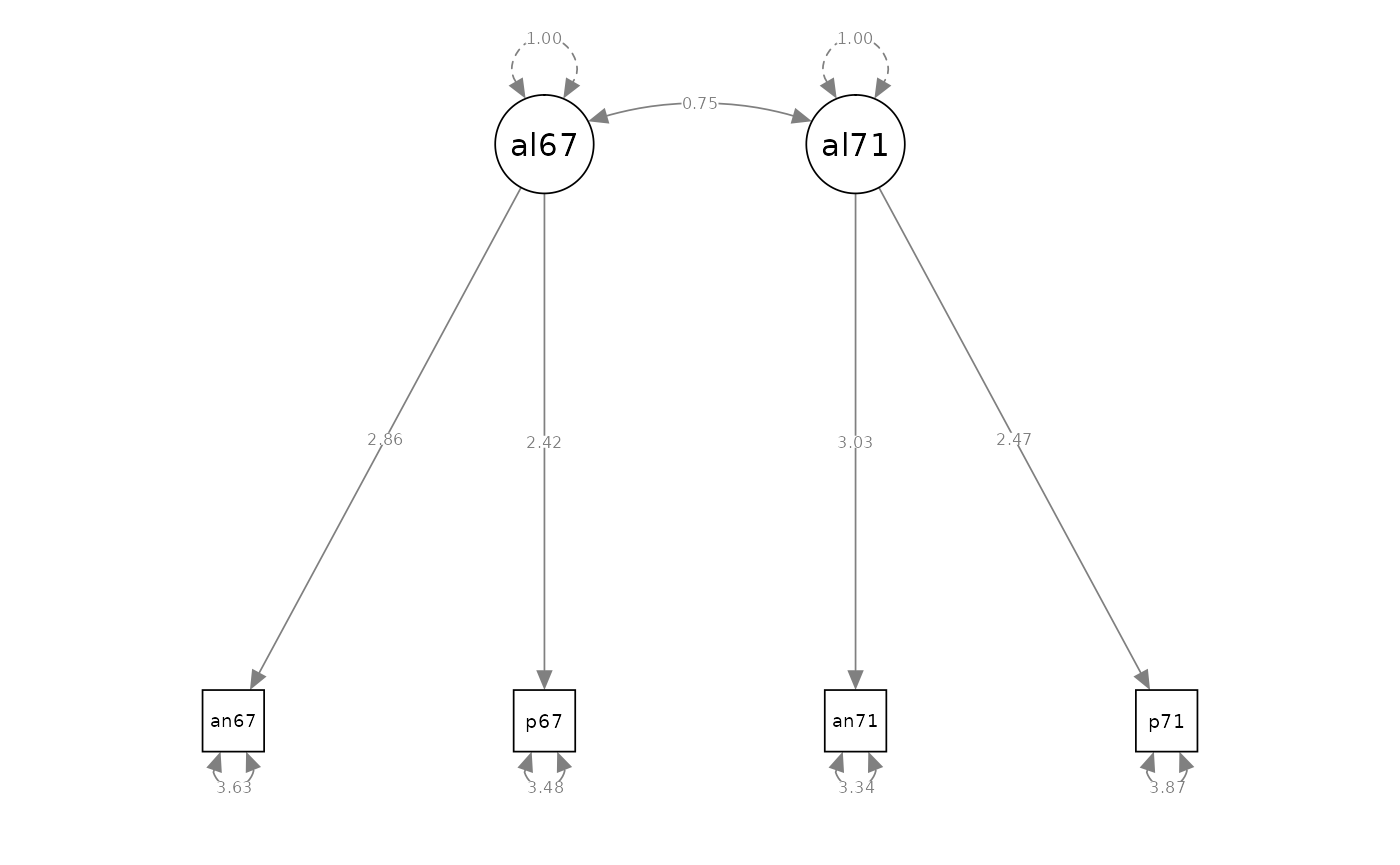
Complete Latent Variable Model
# model specification
model_cfa_6 <- "
# measurement model
alienation67 =~ 1 * anomia67 + powerless67
alienation71 =~ 1 * anomia71 + powerless71
ses =~ 1 * education + sei
# measurement error covariances
anomia67 ~~ anomia71
powerless67 ~~ powerless71
# regression
alienation71 ~ alienation67 + ses
alienation67 ~ ses
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# variances of factors
ses ~~ ses
# residual variances of factors
alienation67 ~~ alienation67
alienation71 ~~ alienation71
# measurement error variances
anomia67 ~~ anomia67
powerless67 ~~ powerless67
anomia71 ~~ anomia71
powerless71 ~~ powerless71
"
# model fitting
fit_cfa_6 <- sem(
model_cfa_6,
sample.cov = alienation,
sample.nobs = 932
)
# results
summary(fit_cfa_6, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 84 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 17
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic 4.735
#> Degrees of freedom 4
#> P-value (Chi-square) 0.316
#>
#> Model Test Baseline Model:
#>
#> Test statistic 2133.722
#> Degrees of freedom 15
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 1.000
#> Tucker-Lewis Index (TLI) 0.999
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -15213.274
#> Loglikelihood unrestricted model (H1) -15210.906
#>
#> Akaike (AIC) 30460.548
#> Bayesian (BIC) 30542.783
#> Sample-size adjusted Bayesian (BIC) 30488.792
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.014
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.053
#> P-value RMSEA <= 0.05 0.930
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.007
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Latent Variables:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 =~
#> anomia67 1.000 2.663 0.774
#> powerless67 0.979 0.062 15.895 0.000 2.606 0.852
#> alienation71 =~
#> anomia71 1.000 2.850 0.805
#> powerless71 0.922 0.059 15.498 0.000 2.628 0.832
#> ses =~
#> education 1.000 2.607 0.842
#> sei 5.219 0.422 12.364 0.000 13.609 0.642
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation71 ~
#> alienation67 0.607 0.051 11.898 0.000 0.567 0.567
#> ses -0.227 0.052 -4.334 0.000 -0.207 -0.207
#> alienation67 ~
#> ses -0.575 0.056 -10.195 0.000 -0.563 -0.563
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .anomia67 ~~
#> .anomia71 1.623 0.314 5.176 0.000 1.623 0.356
#> .powerless67 ~~
#> .powerless71 0.339 0.261 1.298 0.194 0.339 0.121
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> ses 6.798 0.649 10.475 0.000 1.000 1.000
#> .alienation67 4.841 0.467 10.359 0.000 0.683 0.683
#> .alienation71 4.083 0.404 10.104 0.000 0.503 0.503
#> .anomia67 4.731 0.453 10.441 0.000 4.731 0.400
#> .powerless67 2.563 0.403 6.359 0.000 2.563 0.274
#> .anomia71 4.399 0.515 8.542 0.000 4.399 0.351
#> .powerless71 3.070 0.434 7.070 0.000 3.070 0.308
#> .education 2.801 0.507 5.525 0.000 2.801 0.292
#> .sei 264.597 18.126 14.597 0.000 264.597 0.588
semPaths(fit_cfa_6, what = "path", whatLabels = "est", style = "ram")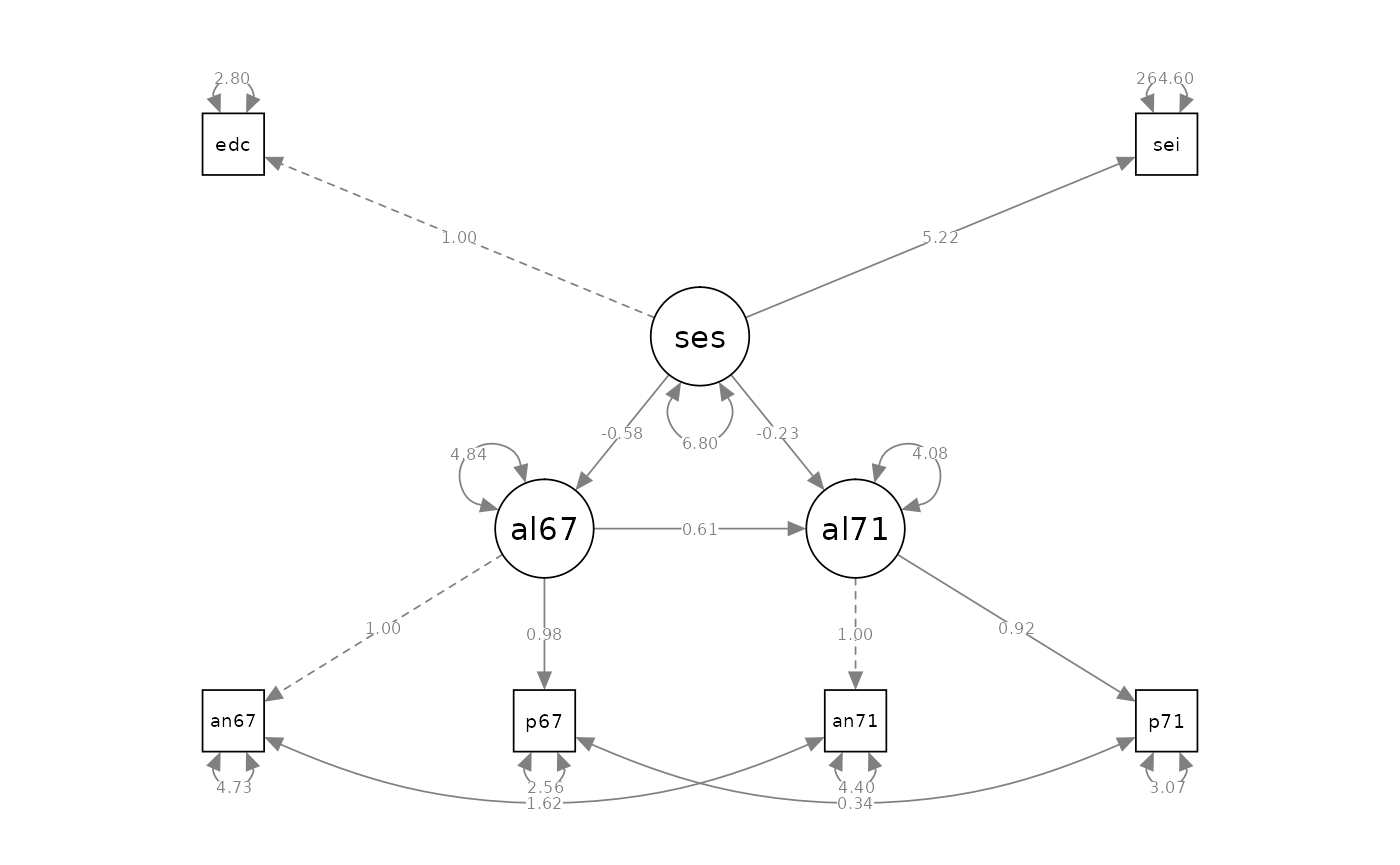
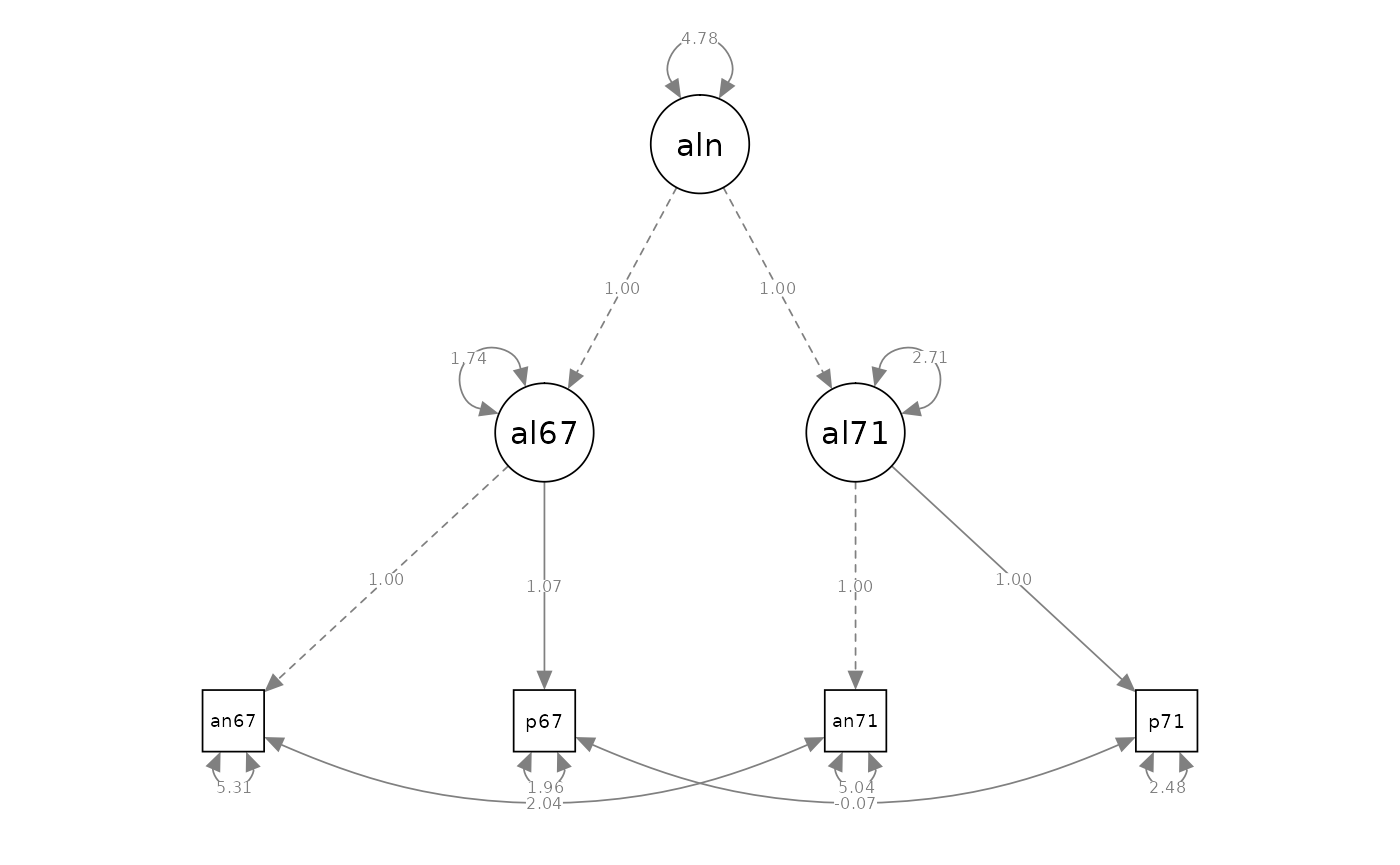
Second-Order Factor Model
# model specification
model_cfa_7 <- "
# measurement model
alienation67 =~ 1 * anomia67 + powerless67
alienation71 =~ 1 * anomia71 + powerless71
alienation =~ a * alienation67 + a * alienation71
# measurement error covariances
anomia67 ~~ anomia71
powerless67 ~~ powerless71
#-------------------------------------------------------
# The syntax above is sufficient to specify this model.
# The syntax below is added to be more explicit
# about the parameters being estimated.
#-------------------------------------------------------
# variance of second-order factor
alienation ~~ alienation
# residual variances of factors
alienation67 ~~ alienation67
alienation71 ~~ alienation71
"
# model fitting
fit_cfa_7 <- sem(
model_cfa_7,
sample.cov = alienation,
sample.nobs = 932
)
#> Warning in lav_model_vcov(lavmodel = lavmodel, lavsamplestats = lavsamplestats, : lavaan WARNING:
#> Could not compute standard errors! The information matrix could
#> not be inverted. This may be a symptom that the model is not
#> identified.
# results
summary(fit_cfa_7, fit.measures = TRUE, standardized = TRUE)
#> lavaan 0.6-7 ended normally after 48 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of free parameters 11
#>
#> Number of observations 932
#>
#> Model Test User Model:
#>
#> Test statistic NA
#> Degrees of freedom -1
#> P-value (Unknown) NA
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) NA
#> Tucker-Lewis Index (TLI) NA
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -8949.380
#> Loglikelihood unrestricted model (H1) -8949.380
#>
#> Akaike (AIC) 17920.761
#> Bayesian (BIC) 17973.971
#> Sample-size adjusted Bayesian (BIC) 17939.036
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA NA
#> 90 Percent confidence interval - lower NA
#> 90 Percent confidence interval - upper NA
#> P-value RMSEA <= 0.05 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Latent Variables:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation67 =~
#> anomia67 1.000 2.553 0.742
#> powerlss67 1.065 NA 2.719 0.889
#> alienation71 =~
#> anomia71 1.000 2.735 0.773
#> powerlss71 1.001 NA 2.737 0.867
#> alienation =~
#> alienatn67 (a) 1.000 0.856 0.856
#> alienatn71 (a) 1.000 0.799 0.799
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .anomia67 ~~
#> .anomia71 2.037 NA 2.037 0.394
#> .powerless67 ~~
#> .powerless71 -0.066 NA -0.066 -0.030
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> alienation 4.775 NA 1.000 1.000
#> .alienation67 1.741 NA 0.267 0.267
#> .alienation71 2.707 NA 0.362 0.362
#> .anomia67 5.305 NA 5.305 0.449
#> .powerless67 1.963 NA 1.963 0.210
#> .anomia71 5.036 NA 5.036 0.402
#> .powerless71 2.484 NA 2.484 0.249
semPaths(fit_cfa_7, what = "path", whatLabels = "est", style = "ram")